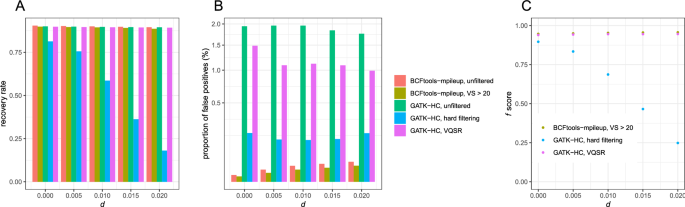
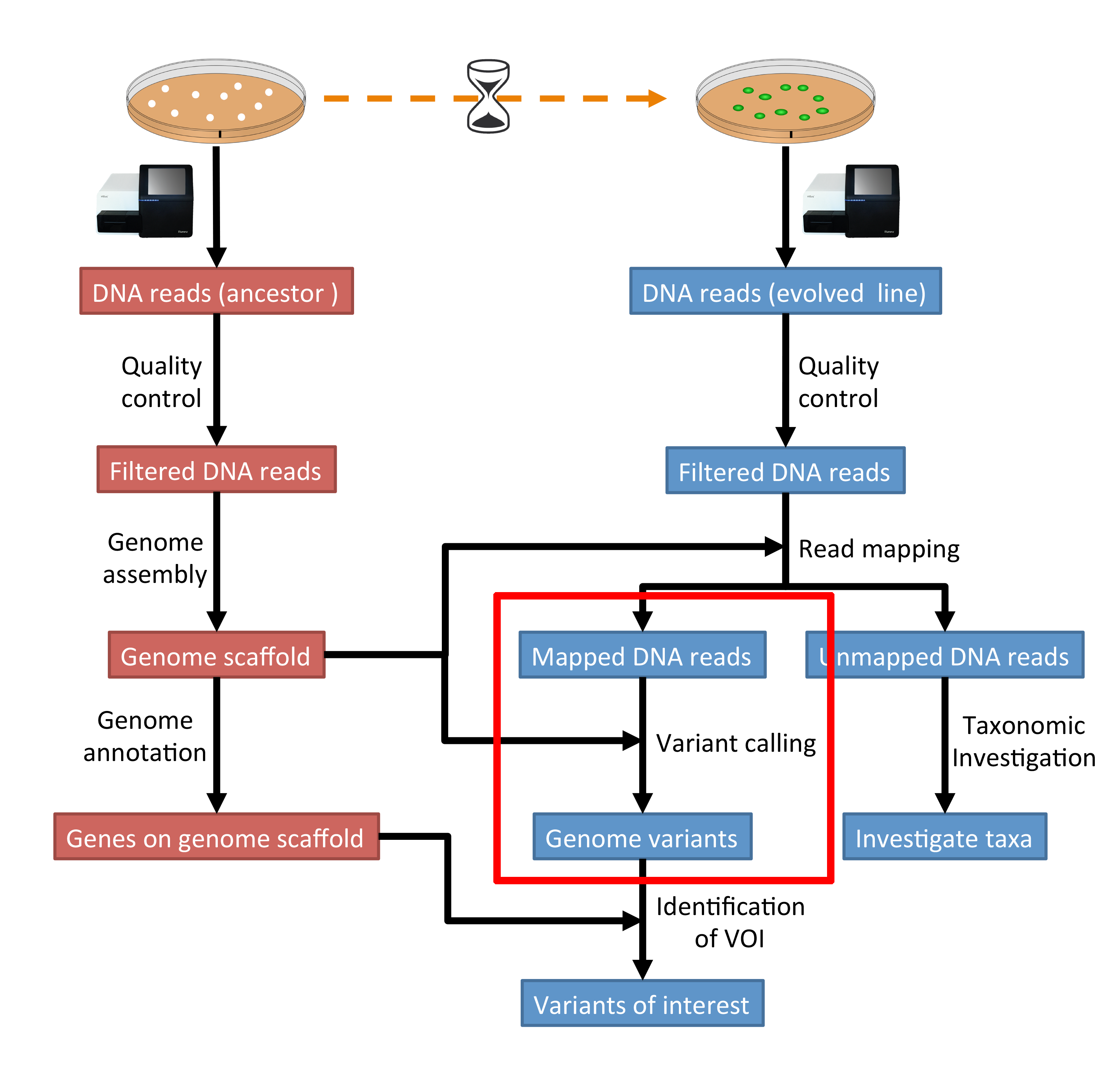
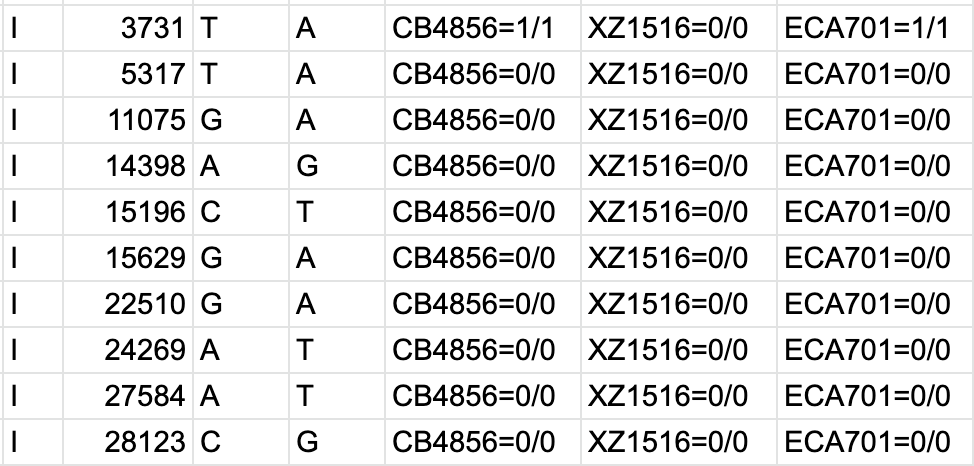
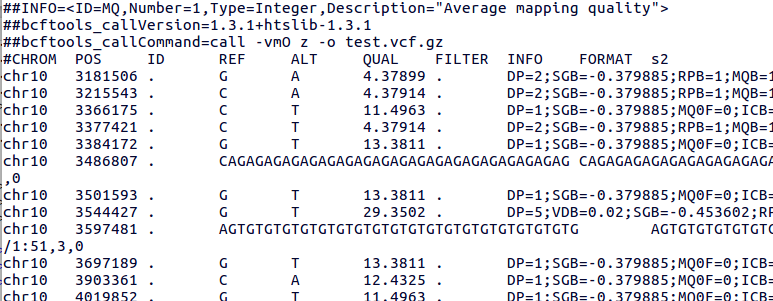
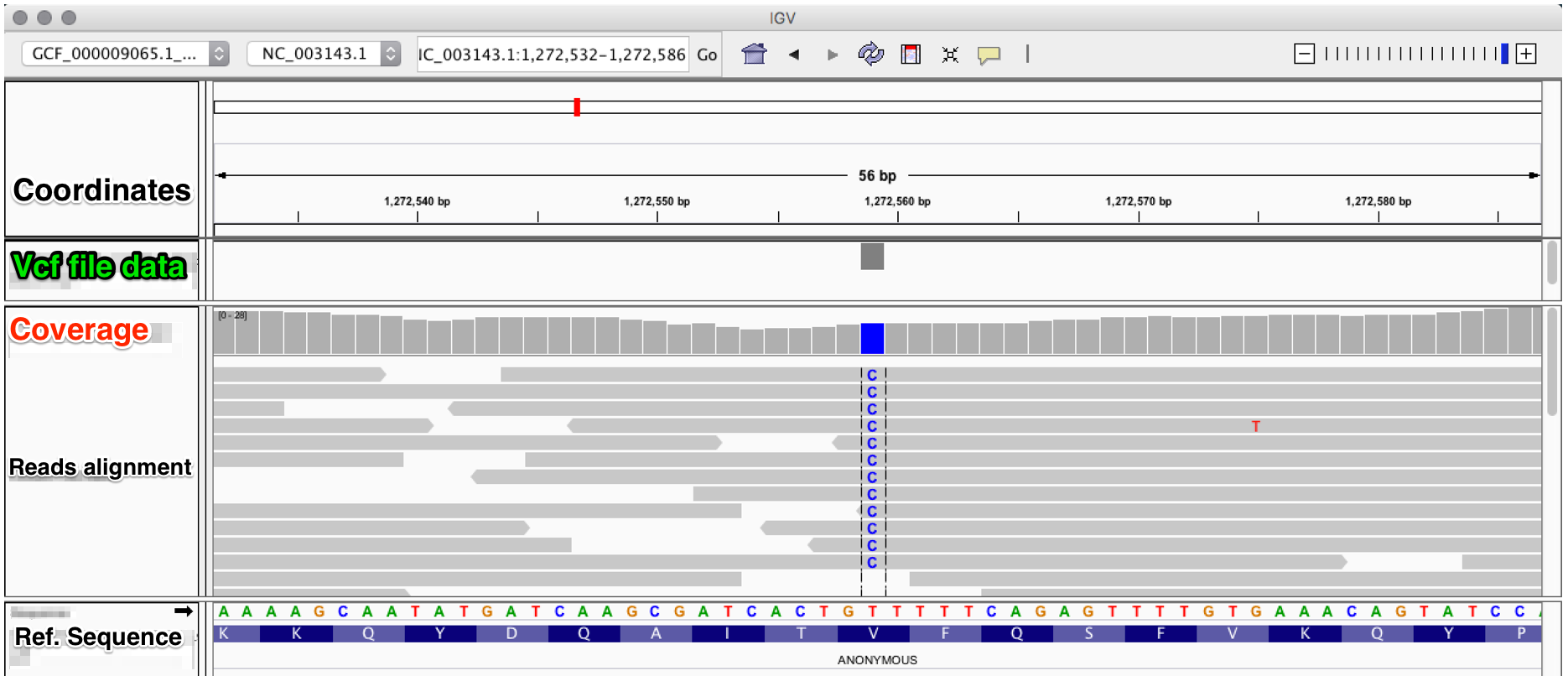
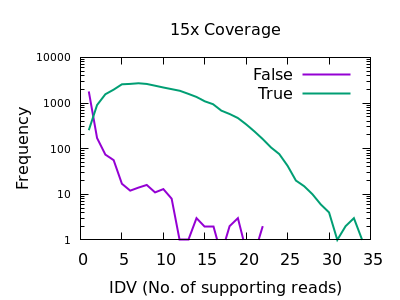
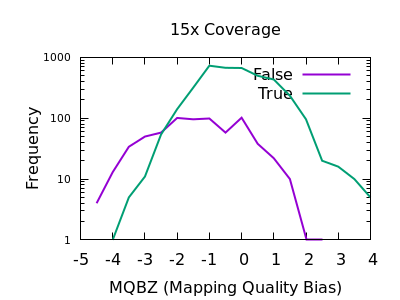
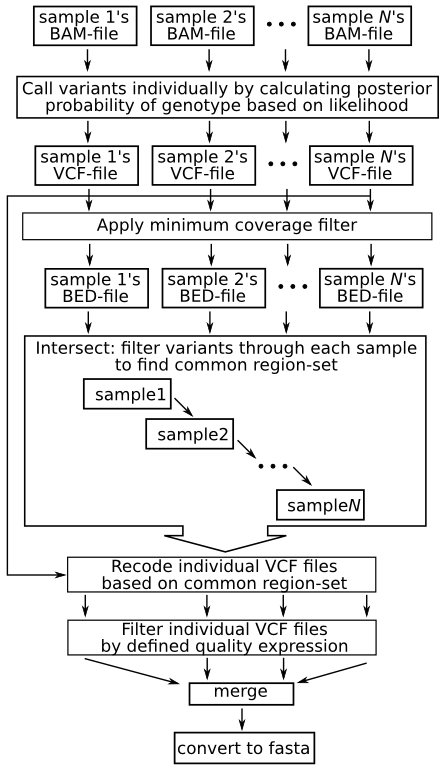
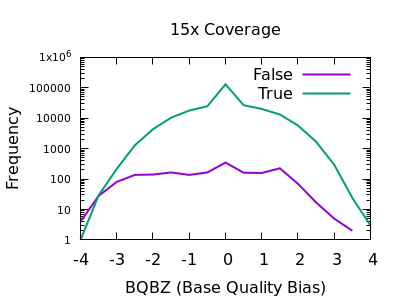
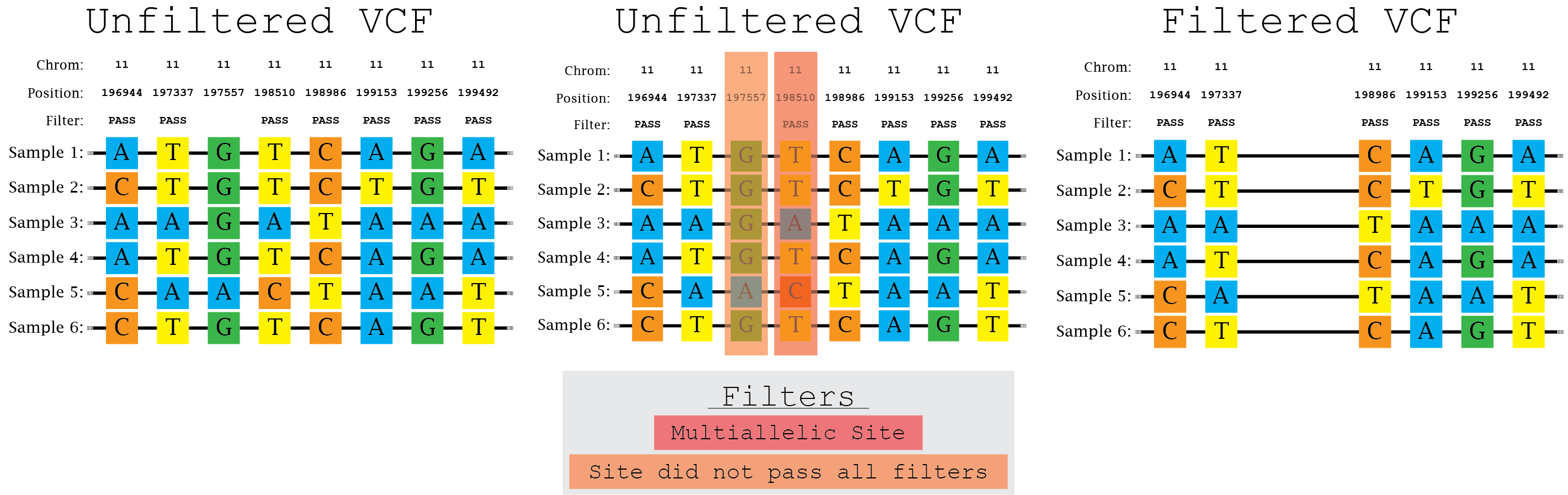
PDF) How to extract and filter SNP data from the genotyping-by- sequencing (GBS) data in vcf format using bcftools

bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube

bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube

bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub
Possible to annotate the FILTER from an annotation VCF file, as an INFO/TAG in the target? · Issue #1187 · samtools/bcftools · GitHub